This guide shows how to perform quality control on a library using the barcoded constructs from the Minigene report and a sample sheet that contains a second, sample-level barcode.
The library itself is generated by ordering the barcoded constructs and then cloning them into the appropriate expression vectors to function as MHC-presented peptides. The sample barcode is then added via PCR primers and few rounds of amplification.
This workflow can be used either on the construct library, the transduced cells, or cells after co-culture. The goal of this step is to confirm that all constructs are present in the approriate proportions. We provide functions to process and analyze the sequencing data.
Preparation
Barcoded construct tables
The Minigene report step of this package provides a table with tiled reference and alternative peptides, along with their respective IDs. When ordering those as gene blocks, one or more construct barcodes are added to the sequence, so we are able to uniquely identify our minigene peptides also when a sequencing read does not reach the mutated base.
Here, we use barcodes from this publication and Github repository:
lib = "https://raw.githubusercontent.com/hawkjo/freebarcodes/master/barcodes/barcodes12-1.txt"
valid_barcodes = readr::read_tsv(lib, col_names=FALSE)$X1These barcodes need to be of the same length and not overlap between samples in one QC run. For the analysis, we manually add them as columns to the peptide/minigene tables:
-
barcode– if only one barcode per construct -
barcode_1,barcode_2, etc. – if multiple barcodes per construct
The other required fields are, for each sample:
-
gene_name– an identifier for the gene, e.g.NRAS -
mut_id– a common identifier forrefandaltsequences, e.g.NRAS_Q61 -
pep_id– a unique identifier for each peptide, e.g.NRAS_Q61L-1 -
pep_type– whether this is areforaltsequence (or other) -
tiled– the tiled nucleotide sequence
For the QC analysis, we will need a named list of these barcoded
Minigene tables. We will usually have assembled them in a
.xlsx file with these barcodes added, however, here we will
use an example dataset:
## normally we'd read all sheets from our barcoded construct file:
# fname = "my_combined_barcoded_file.xlsx"
# sheets = readxl::excel_sheets(fname)
# all_constructs = sapply(sheets, readxl::read_xlsx, path=fname, simplify=FALSE)
all_constructs = example_peptides(valid_barcodes)We can confirm that these barcodes do not overlap:
plot_barcode_overlap(all_constructs, valid_barcodes)
#> Joining with `by = join_by(barcode)`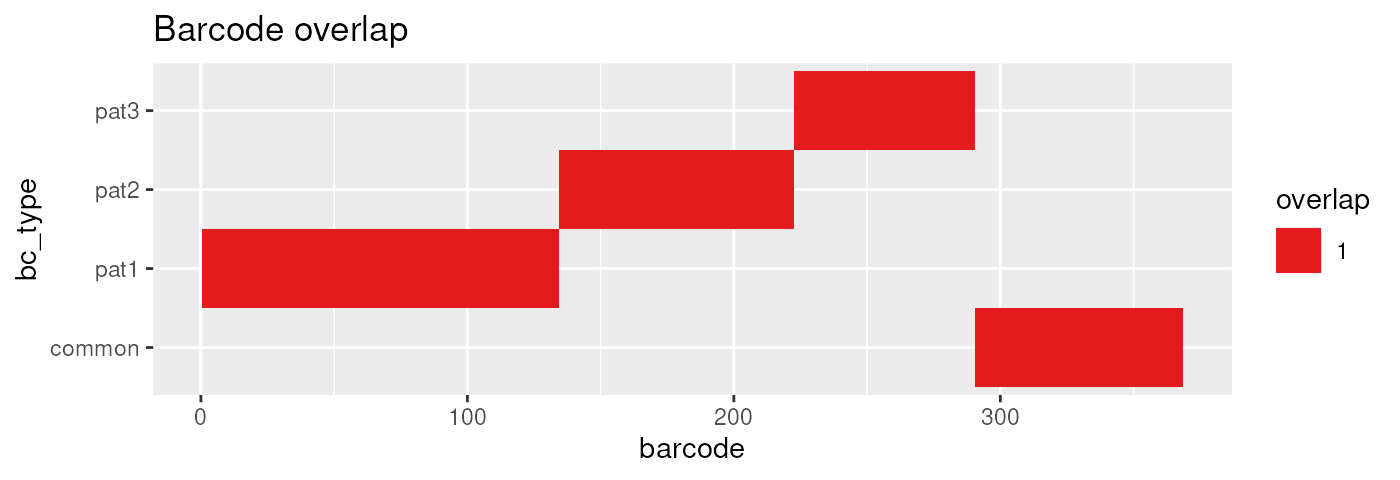
Creating a sample sheet
A sequencing (FASTQ) file will contain a sample barcode, construct barcode, and construct sequence:
fastq_file = "/path/to/your/file.fq.gz"Before we can start working with the FASTQ sequencing output, we need to describe which samples it contains. For this, we create a sample sheet with the following columns:
-
sample_id– a unique identifier of the sample -
patient– the patient or sample group that the sample comes from -
rep– a number indicating the replicate number -
origin– a descriptor of which kind of sample this is -
barcode– the barcode used to label all condition-specific constructs in the sequencing data
For this example, we have already provided a sample sheet with the
package. It includes three samples, two for library QC
(pat2 and pat3) and one with mock-transfected
vs. actual peptide T-cell co-culture (pat1). The
pat1 and pat2 samples additionally include a
common construct set. Whenever we use multiple construct sets in a
sample, we separate them using a +:
sample_sheet = system.file("my_samples.tsv", package="pepitope")Here, we did not actually do any sequencing, so we provide a simulated FASTQ file instead:
fastq_file = example_fastq(sample_sheet, all_constructs)Demultiplexing and counting
Sample demultiplexing
This step is using the fqtk toolkit to split the
multi-sample FASTQ file into individual sample files, separated by their
sample barcode and save to a temporary directory.
Here, read_structures describes how the split these
samples. It has the following possible fields, preceded by the number of
nucleotides in the read. A + instead of a number is used to
indicated to use all remaining nucleotides:
-
B– the sample barcode to split on (required) -
T– the read sequence with the construct barcode and sequence (required) -
S– skip these nucleotides and do not include in output (optional)
In this example, we use a 7-nucleotide barcode for the samples and keep the rest of the read in the split files:
temp_dir = demux_fq(fastq_file, sample_sheet, read_structures="7B+T")
list.files(temp_dir, pattern="\\.fq\\.gz$")
#> [1] "lib1.R1.fq.gz" "lib2.R1.fq.gz" "mock1.R1.fq.gz"
#> [4] "mock2.R1.fq.gz" "screen1.R1.fq.gz" "screen2.R1.fq.gz"
#> [7] "unmatched.R1.fq.gz"You can perform additional quality control steps on the sample-level
FASTQ files, e.g. using fastqc or multiqc (not
included in this package).
Counting construct barcodes
The next step is to count the construct barcodes in each individual
FASTQ file after demultiplexing. Internally, we use the
guide-counter tool to identify the position the construct
barcodes occur in the library reads and subsequently count the number of
occurrences. Here we pass the directory containing the demultiplexed
FASTQ files and the barcode library file with all possible (not only the
used) barcodes:
dset = count_bc(temp_dir, all_constructs, valid_barcodes)
#> Joining with `by = join_by(sample_id)`
#> Joining with `by = join_by(barcode)`Here, dset will be a SummarizedExperiment
object that you can interact with the following way:
-
colData(dset)– access the sample metadata asdata.frame -
rowData(dset)– access the construct metadata asdata.frame -
assay(dset)– access the construct counts asmatrix
Quality Control plots
Plotting total barcode counts
The first overview that we want to get is to know how many barcodes are in which demultiplexed FASTQ, and whether they match the sample we expect them to be from. We can plot this the following way, for total read counts on the left and number of barcodes that have 10 or more reads on the right:
plot_reads(dset)
#> Joining with `by = join_by(barcode)`
#> Joining with `by = join_by(sample_id)`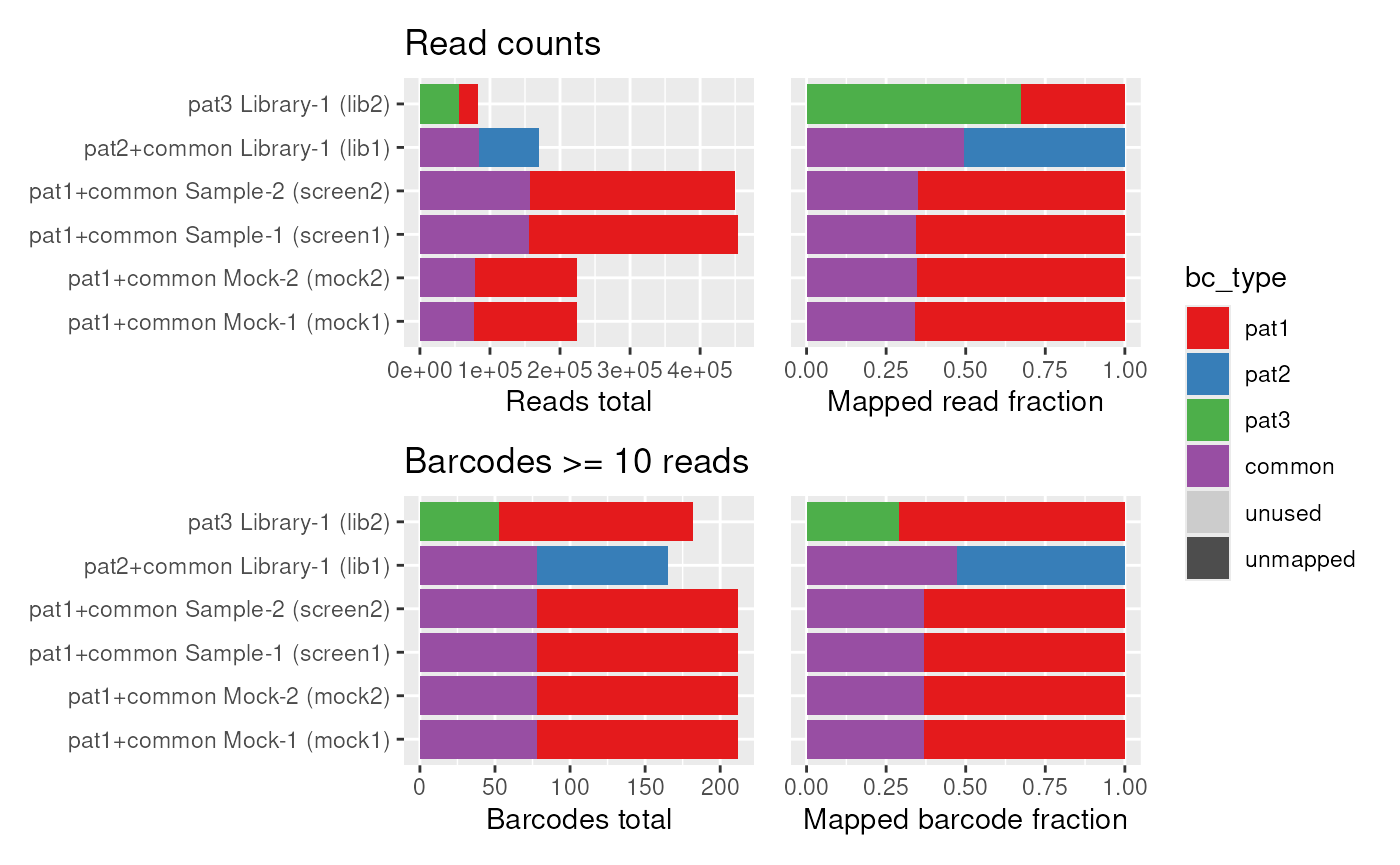
We can also plot this interactively with plotly:
Representation of individual barcodes
The next question we want to ask is whether the individual construct barcodes are equally distributed within a sample, and where any potential contaminations come from. For this, we order the barcodes from least abundant (left) to most abundant (right) and plot a continuous line for how many reads are sequenced of this barcode on the y axis:
plot_distr(dset)
#> Joining with `by = join_by(barcode)`
#> Joining with `by = join_by(sample_id)`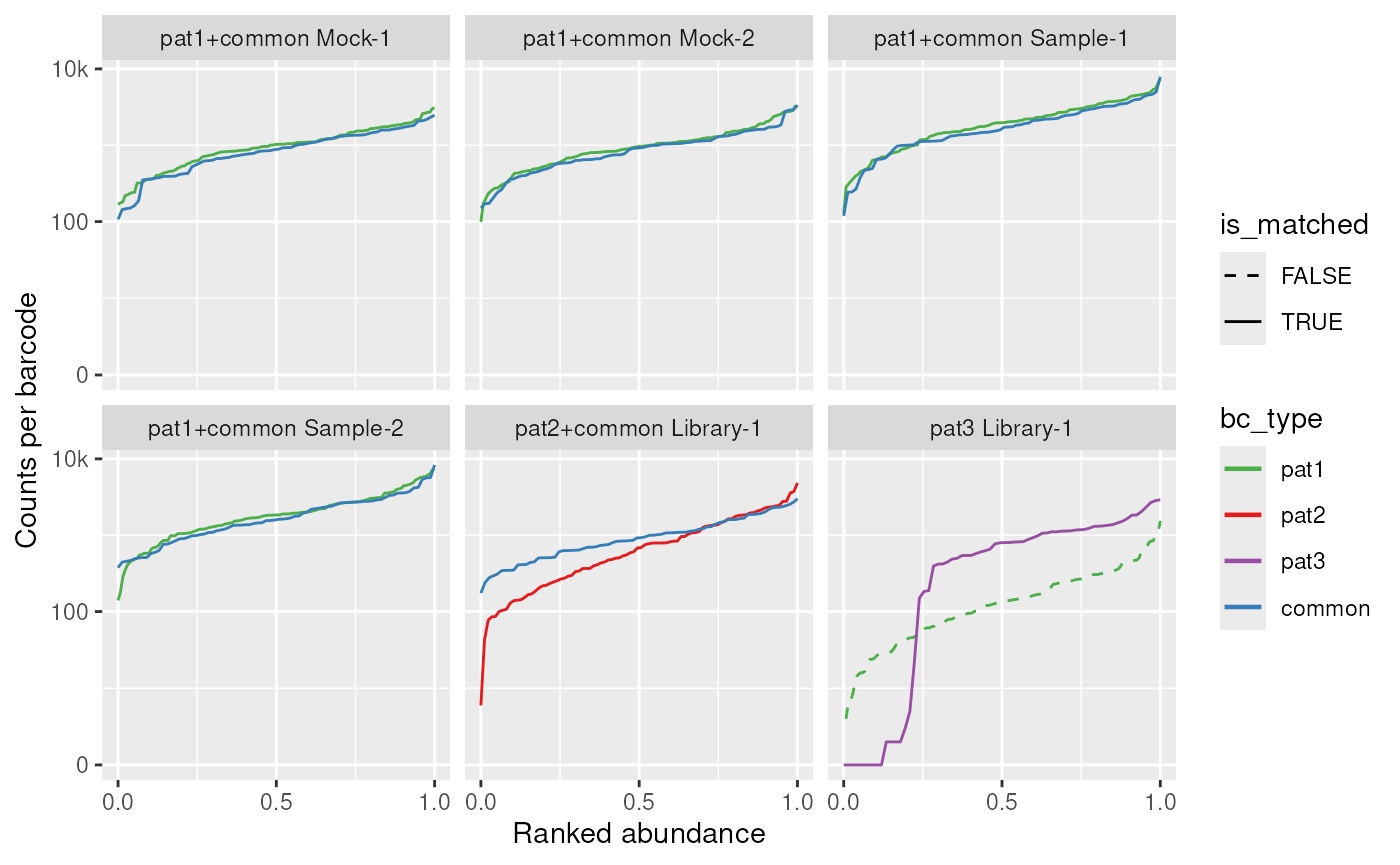
We can make a couple of observations from these plots:
- The
MockandSampleconditions worked well forpat1 - The sample-specific barcodes for
pat2show a high read variance and may be unsuitable for a screen - About a quarter of the sample-specific barcodes for
pat3are lost - There is a sample contamination of
pat1inpat3
We can also plot this interactively with plotly, where
we can hover over with the mouse to see which barcode is in which
position exactly:
library(plotly)
ggplotly(plot_distr(dset), height=500, tooltip="text")
#> Joining with `by = join_by(barcode)`
#> Joining with `by = join_by(sample_id)`